Use of Molecular Tweezers to Study the Effect of Applied Force on σ54 Core-Binding Domain
During the 2011-2012 school year, I wrote my honors undergraduate thesis in Professor Dave Wemmer’s Laboratory at University of California, Berkeley. I performed a molecular tweezers experiment on the transcription initiation protein, σ54, to determine the number of unfolding events in an attempt to better understand the mechanism behind σ54 transcription initiation.
In bacteria, σ factors regulate cellular transcription. They must associate with RNA Polymerase (RNAP) to form the σ-RNAP holoenzyme that then can melt double-stranded DNA and initiate transcription. Most σ factors are part of the σ70 class that can immediately initiate transcription after association with RNA Polymerase. σ54, however, requires activation by an AAA+ ATPase activator protein that remodels the σ54-RNAP complex into an open, active conformation [1].
Currently, it is unclear whether activation works through a single bind-and-release mechanism or whether the ATPase requires multiple rounds to ATP hydrolysis to exert a “tugging” force on σ54. The latter hypothesis is supported by evidence of a fracture point between the two subdomains of the core binding domain of σ54 [2]. My project aimed to mimic the proposed tugging by the ATPase activator to observe the number of protein unfolding events of the core binding domain. Two unfolding events lends further support to the hypothesis that σ54 requires tugging to activate, and the fracture point converts this force into conformational changes to its DNA binding domain that help separate the double-stranded DNA to initiate transcription.
We found that approximately 20% of the core-binding domain of σ54 unfolds before the rest of the domain during force-extension cycles with the molecular tweezers. This was surprising because the two subdomains of σ54 are each around 50% of the domain. Our data suggests that a portion of one of the subdomains is unfolding before the rest. This uncooperative unfolding could be important to the mechanism of σ54 transcription initiation. We are truncating a portion of the domain that is likely to account for this 20% and repeating our molecular tweezers experiment to determine the specific portion of the domain that unfolds before the rest.
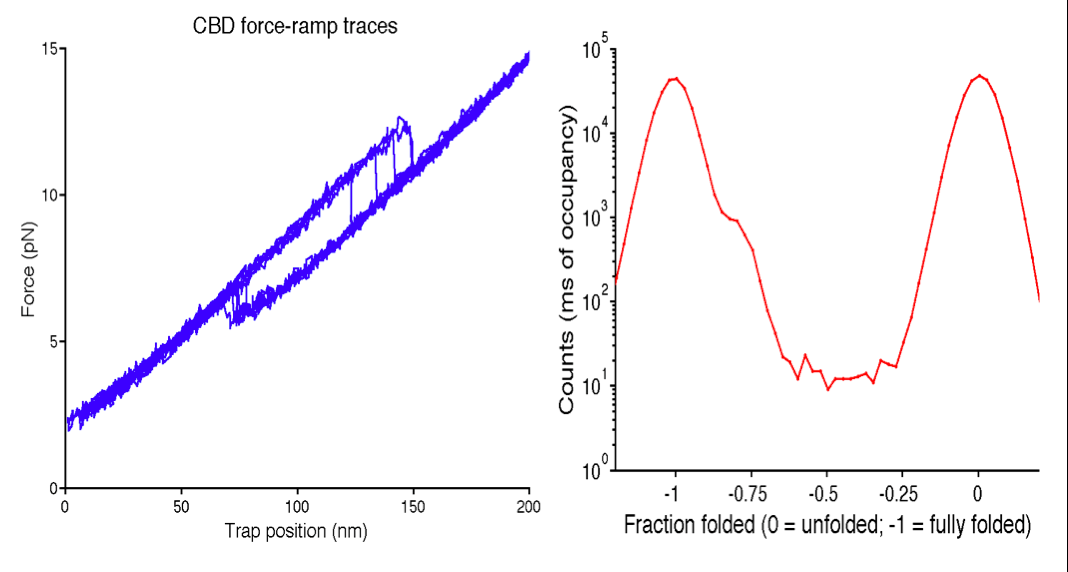
For more information on this project, here are links to my poster and to my undergraduate honors thesis.
-
Wedel, A., Kustu, S. (1995). The bacterial enhancer-binding protein NTRC is a molecular machine: ATP hydrolysis is coupled to transcriptional activation. Genes Dev. 9, 2042-2052.
-
Hong, E., Doucleff, M., Wemmer, D.E. (2009). Structure of the RNA polymerase core-binding domain of sigma54 reveals a likely conformational fracture point. Journal of molecular biology. 390, 70-82.